Spatial Transcriptomics
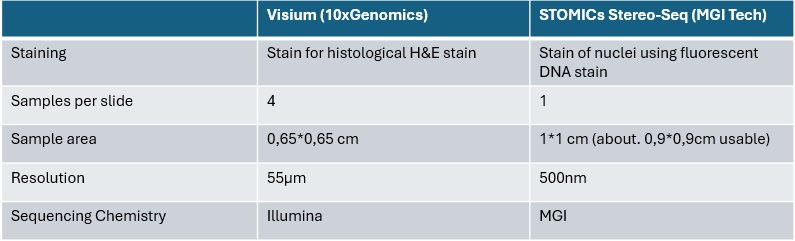
For Spatial transcriptomics, the facility has implemented the Visium workflow of 10xGenomics, as well as the STOMICs Stereo Seq workflow from MGI Tech. Both workflows are based on a tissue kryosection placed onto a chip containing thousands of regularly arranged, area specific barcoded oligonucleotides, that are incorporated into the cDNA generated in this specific area. After staining of the section to visually assign histological areas or single cell nuclei, the tissue is permeabilized so that the mRNAs can bind to the chip surface immediatly below the cell of orgin. Next, a cDNA is generated from all poly-A containing RNAs that incorporates the above mentioned spatial barcode. After sequencing, this barcode allows for annotation of the reads to specific areas in the section allowing for spatially resolved expression profiles down to single cell resolution. The following table gives an overview on the main differences between the two workflows:
More detailed information on the proprietary Visium technology and the Stereo-Seq technology can be found on the respective company´s website (https://www.10xgenomics.com/products/spatial-gene-expression, https://en.stomics.tech/why-stereo-seq/index.html).
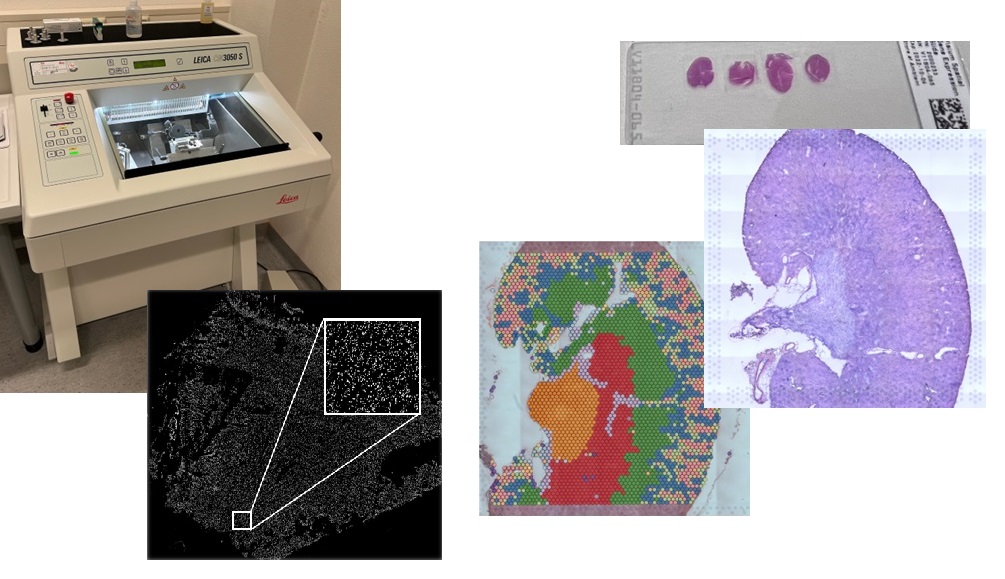
Our Service includes initial consultation concerning project design and sample requirements, generation of images and sequencing libraries from a kryosection, all process quality controls and final sequencing on an Illumina or MGI sequencer (external service). Most recently, our facility is also equipped with a CM3050 S kryostat (Leica) that can be used to generate the kryosections for the spatial transcriptomics workflow.